How To Calculate Sample Size In R
Sample Size Calculations with {pwr}
When designing clinical studies, it is often important to calculate a reasonable approximate of the needed sample size. This is disquisitional for planning, equally you may detect out very quickly that a reasonable study budget and timeline will exist futile. Grant funding agencies volition exist very interested in whether you have a good rationale for your proposed sample size and timeline, then that they can avoid wasting their money.
Fortunately, the {pwr} package helps with many of the needed calculations. Let'due south first install this packet from CRAN, and load it with a library() function.
Remember that you can copy each code chunk to the clipboard, then paste it into your RStudio console (or a script) to edit and run it. Just hover your mouse arrow over the top right corner of each code clamper until a copy icon appears, and then click on it to copy the lawmaking.
install.packages('pwr') library(pwr) The {pwr} package has a number of functions for calculating sample size and power.
| Test | Sample Size Function |
|---|---|
| one-sample, two-sample, and paired t-tests | pwr.t.test() |
| two-sample t-tests (unequal sample sizes) | pwr.t2n.test() |
| two-sample proportion exam (unequal sample sizes) | pwr.2p2n.test() |
| ane-sample proportion test | pwr.p.examination() |
| ii-sample proportion test | pwr.2p.examination() |
| 2-sample proportion examination (diff sample sizes) | pwr.2p2n.examination() |
| 1-fashion balanced ANOVA | pwr.anova.examination() |
| correlation exam | pwr.r.exam() |
| chi-squared test (goodness of fit and association) | pwr.chisq.test() |
| examination for the general linear model | pwr.f2.test() |
Now that you have {pwr} upward and running, we tin can outset with a elementary example.
Sample Size for a Continuous Endpoint (t-test)
Let's propose a written report of a new drug to reduce hemoglobin A1c in type 2 diabetes over a 1 year study period. Yous estimate that your recruited participants will have a mean baseline A1c of nine.0, which volition be unchanged past your placebo, just reduced (on boilerplate) to 7.0 by the study drug.
You need to calculate an consequence size (aka Cohen's d) in order to estimate your sample size. This effect size is equal to the divergence betwixt the means at the endpoint, divided by the pooled standard deviation. Many clinicians tin can judge the ways and the difference, but the pooled standard deviation is not very intutitive. Sometimes yous take an estimate from airplane pilot information (though these tend to take wide confidence intervals, as airplane pilot studies are small). In other circumstances, y'all can estimate a standard deviation for Hgb A1c from values from a large data warehouse. When you don't have either of these, it tin be helpful to first by estimating the range of values.
This is something that is intutitive, and experienced clinicians tin do fairly easily. But become a few clinicians in a room, and ask them for the highest and lowest values of HgbA1c that they take ever seen. You will rapidly find a minimum and maximum that y'all can estimate as the range (in this instance, permit'due south say 5.0 and 17.0 for min and max of Hgb A1c). This range divided by four is a reasonable crude estimate of the standard departure. Remember that a normally distributed continuous value volition accept a 95% confidence interval that is plus or minus 1.96 standard deviations from the sample mean. Round this upwards to 2 for the total range, and you can see why we divide the range by 4 to get an estimate of the standard deviation.
In our case, the divergence is 2 and the range/four (estimate of SD) is 3. So our effect size (Cohen's d) is 0.66. Plug in 0.66 for d in the code chunk beneath, and run this code clamper to go an approximate of the n in each arm of a ii armed study with a two sample t-test of the master endpoint.
pwr:: pwr.t.examination(n = NULL, sig.level = 0.05, blazon = "two.sample", alternative = "ii.sided", power = 0.fourscore, d = __) Nosotros come up up with 37.02 participants in each grouping to provide fourscore% power to notice a departure of two in HgbA1c, bold a standard departure of 3, using a two-sided alpha of 0.05. To conduct this written report, assuming a 20% dropout rate in each arm, would crave 37+eight subjects per arm, or 90 overall. At an enrollment rate of 10 per month, it will require 9 months to enroll all the participants, and 21 months (ix + 12 calendar month intervention) to complete the data collection.
It is a very common mistake to look at the result for northward, and assume that this is your total sample size needed. The n provided past {pwr} is the number per arm. You lot need to multiply this northward by the number of artillery (or in a paired assay, by the number of pairs) to go your total n.
Another mutual mistake is to assume no dropout of participants. It is important to have a reasonable estimate (x-20% for brusk studies, 30-50% for long or demanding studies) and inflate your intended sample size by this amount. It is even better if you know from like studies what the actual dropout rate was, and use this every bit an estimate (if in that location are similar previous studies). Every bit a general rule, information technology is amend to exist conservative, and estimate a larger sample size, than to end up with p = 0.07.
Once you define your test type (the options are "two.sample", "one.sample", and "paired"), and the alternative ("2.sided", "greater", or "less"), four variables remain in a sample size and ability adding.
These are the remaining four arguments of the pwr.t.test() function. These are:
-
n
-
the significance level (sig.level)
-
the ability
-
the effect size (Cohen's d)
If you lot know any three of these, y'all tin calculate the fourth. In order to do this, you ready the ane of these four arguments that you lot want to calculate equal to Nada, and specify the other 3. Imagine that nosotros only have plenty funds to run this report on 50 participants. What would our power be to detect a divergence of ii in Hgb A1c? You tin can set the power to NULL, and the n to 25 (remember that due north is per arm), and run the clamper below.
pwr:: pwr.t.examination(n = __, # notation that due north is per arm sig.level = 0.05, type = "ii.sample", alternative = "two.sided", power = __, d = 0.66) We finish up with 62.8% ability, assuming no participant dropout (which is an extremely unlikely assumption). You tin can do the same thing, changing the NULL, to calculate an effect size or a significance level, if you have any need to.
In nearly cases, you are calculating a sample size, then realizing that you might not accept that much money/resources. Then many calculate the ability yous would obtain given the resources you actually take. Let's testify a few more examples.
One Sample t-examination for Lowering Creatinine
Eddie Enema, holistic healer, has proposed an unblinded pilot report of thrice-daily 2 liter enemas with "Eddie'south Dialysis Cleanser," a proprietary mix of vitamins and minerals, which he believes will lower the serum creatinine of patients on the kidney transplant waiting list past more than 1.0 1000/dL in 24 hours. The creatinine SD in this grouping of patients is 2.
The zero hypothesis is < i.0 g/dL. The culling hypothesis is >= 1.0 chiliad/dL.
Cohen'southward d is one.0 (the proposed modify, or delta)/2 (the SD) = 0.5.
How many participants would Eddie Enema have to recruit to take 80% power to exam this one-sample, 1-sided hypothesis, with an alpha of 0.05?
Bank check each of the argument values and run the chunk beneath to find out.
pwr:: pwr.t.test(northward = Cipher, # annotation that n is per arm sig.level = 0.05, type = "ane.sample", alternative = "greater", ability = 0.8, d = 0.five) Fast Eddie would demand to recruit slightly more than 26 participants (you always take to round up to get whole human participants) to have 80% ability, assuming no dropout between the first and third enema, or before the blood depict 24 hours after baseline. Note that since this is an unblinded, i-sample study, The due north in the results is multiplied by the number of arms (there is merely 1 arm) to requite you lot a sample size of 27.
A notation near alpha and beta
alpha is described as the type I error, or the probability of declaring significant a difference that is non truly significant. We frequently use a 2-sided alpha, which cuts the region of significance in half, and distributes it to both tails of the distribution, assuasive for both significant positive and negative differences. Alpha is unremarkably set at 0.05, which works out to 0.025 on each tail of the distribution with a two-tailed alpha.
beta is the power, or (1- the gamble of type Ii error). Type Two error is the probability of missing a significant result and declaring it nonsignificant after hypothesis testing. Power is often ready at lxxx%, or 0.viii, just tin exist 90%, 95%, or 99%, depending on how of import it is non to miss a significant event, and how much money and time you accept to spend (both of which tend to increment N and power).
There is often an important tradeoff between type I and type 2 error. Things that decrease blazon Ii error (increase ability) like spending more fourth dimension and money for a larger N, will increase your risk of type I error. Conversely, reducing your risk of type I error volition generally increase your adventure of type Two error. You may be in situations in which you have to make up one's mind which type of error is more of import to avert for your clinical situation to maximize do good and minimize harms for patients.
Paired t-tests (before vs after, or truly paired)
Every bit you can see from the higher up case, you can utilise a before-after blueprint to measure differences from baseline, and substantially catechumen a ii-sample paired design (each participant's baseline measurement is paired with their post-intervention measurement) to a single sample pattern based on the deviation between the earlier and after values.
The earlier-afterward (or baseline-postintervention) blueprint is probably the most common paired design, only occasionally we have truly paired designs, like when we exam an ointment for psoriasis on 1 arm, and use a placebo or sham ointment on the other arm. When this is possible, through bilateral symmetry (this also works for eyedrops in eyes, or dental treatments), it is much more efficient (in the recruiting sense) than recruiting divide groups for the treatment and control artillery.
To see the difference between two-sample and paired designs, run the lawmaking clamper below, for a two-sample written report with a Cohen's d of 0.8 and eighty% power. Then change the type to "paired", and see the outcome on sample size.
pwr:: pwr.t.test(n = Cypher, # note that n is per arm sig.level = 0.05, type = "ii.sample", alternative = "two.sided", power = 0.8, d = 0.8) Annotation that this changes the needed sample size from 52 subjects (26 per arm) to xv subjects (as there is only one participant needed each paired awarding of 2 written report treatments, and northward in this case indicates the number of pairs), though it would be wise to randomize patients to having the handling on the right vs. left arm (to maintain the blind). This is a large gain in recruiting efficiency. Use paired designs whenever you tin.
two Sample t tests with Unequal Study Arm Sizes
Occasionally investigators want unbalanced arms, as they feel that patients are more likely to participate if they accept a greater chance of receiving the study drug than the placebo. It is fairly common to use ii:1 or 3:ane ratios. Larger ratios, like 4:1 or v:one, are thought to hazard increasing the placebo response rate, every bit participants assume that they are on the active drug. This is somewhat less efficient in recruiting terms, simply it may amend the recruiting rate enough to recoup for the loss in efficiency.
This requires a slightly different function, the pwr.t2n.exam() office. Allow's look at an instance below. Instead of north, we have n1 and n2, and nosotros have to specify one of these, and leave the other equally Cipher. Or we can attempt a variety of ratios of n1 and n2, leaving the power set to NULL, and test numbers to produce the desired ability.
Nosotros are proposing a study in which the expected reduction in systolic blood pressure is 10 mm Hg, with a standard departure of twenty mm Hg. We choose an n1 of 40, and a power of fourscore%, and then permit the role determine n2.
pwr:: pwr.t2n.test(n1 = forty, n2 = Nil, sig.level = 0.05, alternative = "ii.sided", power = 0.8, d = 0.5) In this example, n2 works out to slightly over 153 in the drug arm, or about 4:1.
Calculating effect size, or Cohen's d.
You tin calculate the d value yourself. Or, you tin brand your life easier to permit the {pwr} parcel practise this for you. You can leave the calculation of the delta/SD to the program, by setting
d = (twenty-10)/20, and the program will calculate the d of 0.v for yous.
Nosotros can also round upwardly the ratio to 4:1 (160:twoscore) and determine the resulting power.
pwr:: pwr.t2n.examination(n1 = c(40), n2 = c(160), sig.level = 0.05, alternative = "two.sided", power = NULL, d = 0.5) This provides a ability of 80.3%.
Testing Multiple Options and Plotting Results
It tin can be helpful to compare multiple scenarios, varying the n or the estimated effect size, to examine merchandise-offs and potential scenarios when planning a trial. Y'all can examination multiple particular scenarios by listing the variables in a concatenated vector, as shown below for n1 and n2.
pwr:: pwr.t2n.test(n1 = c(40, sixty, fourscore), n2 = c(fourscore, 120, 160), sig.level = 0.05, culling = "2.sided", power = NULL, d = 0.5) This provides 3 distinct scenarios, with iii pairs of n1/n2 values, and the calculated ability for each scenario.
You can also examine many scenarios, with the sequence role, seq(). For the sequence function, three arguments are needed:
-
from, the number the sequence starts from
-
to, the number the sequence ends at (inclusive)
-
past, the number to increment by
Notation that the length of the sequences produced by seq() must match (or exist a multiple of the other) if you are sequencing multiple arguments, and then that in that location is a number for each scenario. If the lengths of the sequences are multiples of each other (viii and 4 in the instance below), the shorter sequence (n2) will exist silently "recycled" (used once more in the same guild) to produce a vector of matching length (8).
pwr:: pwr.t2n.test(n1 = seq(from = 40, to = 75, by = v), n2 = seq(threescore, 120, 20), sig.level = 0.05, alternative = "two.sided", power = NULL, d = 0.5) Sometimes information technology is helpful to look at multiple scenarios and plot the results. You can do this past leaving n = NULL, and plotting the results, as seen below. The nil value volition exist varied across a reasonable range, and the results plotted, with an optimal value identified. The plot function will utilise ggplot2 if this package is loaded, or base R plotting if ggplot2 is not available. As you lot tin can see below, you can alter the ggplot2 plot of the results with standard ggplot2 functions.
results <- pwr:: pwr.t2n.test(n1 = c(40), n2 = Zilch, sig.level = 0.05, alternative = "two.sided", power = 0.fourscore, d = 0.5) plot(results) + ggplot2:: theme_minimal(base_size = 14) + labs(title = 'Optimizing Sample Size for my two-Sided t examination', subtitle = "Ever Round up for Whole Participants, N = 194") 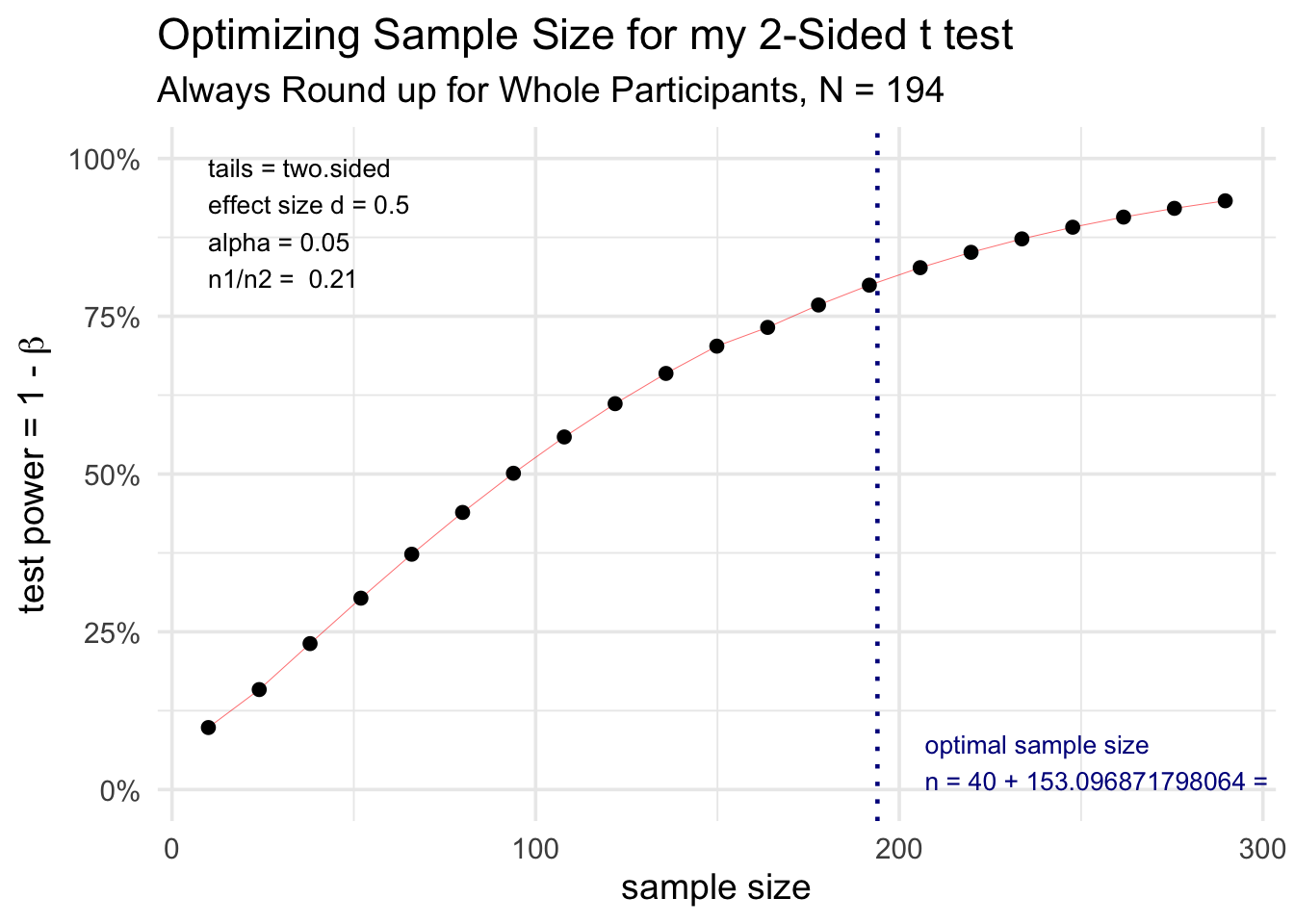
Notation that the results object is a listing, and you can access private pieces with the dollar sign operator, so that `results$n1` equals 40, and `results$n2` equals 153.
You can examine the components of the results object in the Environment pane in RStudio. You can use these in inline R expressions in an Rmarkdown document to write up your results. Retrieve that each inline R expression is wrapped in backwards apostrophes, like `r code` (using the graphic symbol to the left of the i key on the standard United states of america keyboard), and starts with an r to let the computer know that the incoming lawmaking is written in R. This helps you write upward a sentence like the below for a grant application:
Using an estimated effect size of 0.v, with a two-sided alpha of 0.05, we calculated that for 40 participants in group 1, 153.0968718 participants would be needed in group 2 to produce a ability of 0.eight.
When y'all knit an Rmarkdown file with these inline R expressions, each will be automatically converted to the result number and appear every bit standard text.
Your Turn
Endeavor computing the sample size or power needed in the continuous outcome scenarios below. See if yous tin plot the results equally directed by editing the code chunks.
Scenario 1: FEV1 in COPD
You want to increase the FEV1 (forced expiratory volume in 1 second) of patients with COPD (chronic obstructive pulmonary illness) past ten% of predicted from baseline using weekly inhaled stem cells vs. placebo. Unfortunately, the standard divergence of FEV1 measurements is twenty%. You want to have fourscore% power to discover this difference, with a 2-sided alpha of 0.05, with equal north in each of the two artillery. Fill in the blanks in the code chunk below to calculate the sample size needed (n ten number of artillery). Recollect that the effect size (Cohen's d) = change in endpoint (delta)/SD of the endpoint.
pwr:: pwr.t.test(northward = __, # note that n is per arm sig.level = __, blazon = "__", culling = "__", power = __, d = __) You should get 128 participants (bold no dropout) from 64 per arm. Cohen's d is x/20 = 0.5. It can be catchy to keep the blazon of "two.sample" and the culling of "two.sided" straight. Just you lot can practice this!
Scenario two: BNP in CHF
You lot want to subtract the BNP (encephalon natriuretic poly peptide) of patients with CHF (congestive centre failure) by 300 pg/mL from baseline with a new oral intropic agent vs. placebo. BNP levels go up during worsening of centre failure, and a variety of effective treatments lower BNP, which can function as surrogate mark in clinical trials. The standard difference of BNP measurements is estimated at 350 pg/mL. You want to have eighty% ability to detect this divergence, with a 2-sided alpha of 0.05, with equal northward in each of the two arms. Also consider an culling scenario with a change in BNP of only 150 pg/mL. Remember that the issue size (Cohen's d) = modify in endpoint (delta)/SD of the endpoint. Fill in the blanks in the code chunk below (two scenarios) to calculate the sample size needed (due north x number of arms) for both alternatives.
pwr:: pwr.t.test(n = __, # annotation that n is per arm sig.level = __, type = "ii.__", alternative = "two.__", power = __, d = __/__) pwr:: pwr.t.test(due north = __, # note that n is per arm sig.level = __, type = "2.__", alternative = "two.__", power = __, d = __/__) You should get 46 participants (assuming no dropout) from 23 per arm x 2 artillery, or 174 participants (87x2) with the alternative effect size. The effect size (Cohen'south d) is 300/350 = 0.86 in the original, and 150/350 (0.43) in the alternative effect.
Note that you can allow R summate the Cohen's d - but type in 300/350 and 150/350, and R volition use these as values of d.
Scenario 3: Barthel Index in Stroke
You want to increment the Barthel Activities of Daily Living Index of patients with stroke past 25 points from baseline with an intensive in-home PT and OPT intervention vs. usual intendance (which usually increases BADLI past only 5 points). You roughly estimate the standard divergence of Barthel alphabetize measurements as 38. You desire to have 80% power to detect this deviation, with a 2-sided alpha of 0.05, with equal n in each of the two artillery. You want to consider multiple possible options for n, and plot these for a dainty figure in your grant application. Fill in the blanks in the code chunk below to calculate and plot the sample size needed (n x number of arms).
results <- pwr:: pwr.t.examination(north = __, # note that n is per arm sig.level = __, blazon = "two.__", culling = "ii.sided", power = __, d = __ ) plot(results) You should get an optimal sample size of 116 participants (assuming no dropout) from 58 per arm x two artillery, with a dainty plot to evidence this in your grant proposal. The event size (Cohen's d) is (25-5)/38 = 0.526.
Sample Sizes for Proportions
Let's presume that patients discharged from your hospital after a myocardial infarction take historically received a prescription for aspirin 80% of the fourth dimension. A nursing quality improvement project on the cardiac flooring has tried to increment this rate to 95%. How many patients do you lot need to rail later the QI intervention to decide if the proportion has truly increased?
-
the naught hypothesis is that the proportion is 0.8
-
the alternative hypothesis is that the proportion is 0.95.
For this, nosotros need the pwr.p.test() function for one proportion.
We will also use a congenital-in role of {pwr}, the ES.h() part, to help usa calculate the outcome size. This function takes our 2 hypothesized proportions and calculates an effect size with an arcsine transformation.
pwr.p.examination(h = ES.h(p1 = 0.95, p2 = 0.lxxx), northward = Nil, sig.level = 0.05, power = 0.80, alternative = "greater") ## ## proportion power calculation for binomial distribution (arcsine transformation) ## ## h = 0.4762684 ## n = 27.25616 ## sig.level = 0.05 ## power = 0.8 ## alternative = greater Nosotros need to evaluate at least the next 28 patients discharged with MIs to accept 80% ability to test this one-sided hypothesis.
A note about test sided-ness and publication.
Frequently in common use, yous may only exist focused on an increase or decrease in a proportion or a continuous effect, and a one-sided test seems reasonable. This is fine for internal use or local quality comeback work.
Still, for FDA approval of a drug, for grant applications, or for periodical publications, the standard is to ever employ 2-sided tests, being open up to the possibility of both improvement or worsening of the outcome you are studying. This is important to know before you submit a grant application, a manuscript for publication, or a dossier for FDA blessing of a drug or device.
Sample size for ii proportions, equal n
For this, we need the pwr.2p.test() office for two proportions.
You want to calculate the sample size for a study of a cardiac plexus parasympathetic nervus stimulator for pulmonary hypertension. Yous expect the baseline one year mortality to be 15% in loftier-risk patients, and expect to reduce this to 5% with this intervention. You will compare a sham (turned off) stimulator to an agile stimulator in a ii arm study. Use a 2-sided blastoff of 0.05 and a power of 80%. Re-create and edit the code chunk below to determine the sample size (due north, rounded up) per arm, and the overall sample size (2n) fo the report.
pwr.p.test(h = ES.h(p1 = __, p2 = __), n = __, sig.level = __, ability = __, alternative = "__") We need to enroll at least 67 per arm, or 134 overall.
Dichotomous endpoints are generally regarded every bit having greater clinical significance than continuous endpoints, but often crave more ability and sample size (and more than money and time). Most investigators are short on money and time, and adopt continuous outcome endpoints.
Sample size for 2 proportions, unequal arms
For this, we need the pwr.2p2n.test() role for 2 proportions with diff sample sizes.
Imagine yous want to enroll class IV CHF patients in a device trial in which they will be randomized three:1 to a device (vs sham) that restores their serum sodium to 140 mmol/L and their albumin to 40 mg/dL each night. You await to reduce i yr bloodshed from 80% to 65% with this device. Y'all desire to know what your ability will be if you enroll 300 in the device arm and 100 in the sham arm.
pwr.2p2n.test(h = ES.h(p1 = __, p2 = __), n1 = __, n2 = __, sig.level = __, power = __, alternative = "two.sided") This (300:100) enrollment will have 84.half dozen% ability to detect a change from xv% to 5% mortality, with a 2-sided blastoff of 0.05.
Your Turn
Try calculating the sample size or power needed in the proportional outcome scenarios below. Encounter if y'all can plot the results as directed by editing the code chunks.
Scenario 1: Mortality on Renal Dialysis
Yous desire to subtract the mortality of patients on renal dialysis, which averages xx% per year in your local dialysis center. Yous volition randomize patients to a parcel of statin, aspirin, beta blocker, and weekly erythropoietin vs. usual intendance, and hope to reduce almanac bloodshed to ten%. You want to take 80% power to detect this deviation, with a 2-sided blastoff of 0.05, with equal n in each of the two arms. Make full in the blanks in the lawmaking chunk beneath to summate the sample size needed (n x number of arms).
pwr.p.exam(h = ES.h(p1 = __, p2 = __), north = __, sig.level = .05, power = __, alternative = "two.__") You always circular up outset (to whole participants per arm), then multiply by the number of arms. You will need a minimum of 98 per arm, for a total of 196 participants needed to complete the trial.
Scenario 2: Intestinal anastomosis in Crohn'due south illness
You want to subtract 1-yr endoscopic recurrence charge per unit in Crohn'south disease from 90% to 70%. A local surgeon claims that his new "slipknot anastomosis" technique volition accomplish this, by reducing colonic backwash and thereby, reducing endoscopic recurrence. You want to have eighty% power to notice this difference, with a ii-sided alpha of 0.05, with equal n in each of the 2 artillery. Also consider an alternative, more than conservative scenario with a endoscopic recurrence rate of 80% with the new method. Fill in the blanks in the lawmaking chunk below to calculate the sample size needed (n x number of arms) for both alternatives.
pwr.p.exam(h = c(ES.h(p1 = 0.9, p2 = __)), n = NULL, sig.level = .05, power = __, culling = "two.sided") pwr.p.examination(h = c(ES.h(p1 = __, p2 = __)), north = __, sig.level = __, ability = .80, alternative = __) With the originally claimed recurrence proportion of 70%, you will need 30 participants per arm, or 60 for the whole study. The more than bourgeois estimate will crave 98 subjects per arm, or 196 for the whole study.
Scenario 3: Metformin in Donuts
Your local endocrinologist has identified consumption of glazed donuts as a major risk cistron for evolution of type 2 diabetes in your region. She proposes to randomize participants to glazed donuts spiked with metformin vs usual donuts, expecting to reduce the one year proportion of prediabetics with a HgbA1c > vii.0 from 25% to 10%. Yous want to have fourscore% power to observe this difference, with a 2-sided alpha of 0.05, with 2 times as many participants in the metformin donut arm. Yous desire to consider multiple possible sample sizes (n = 25, 50, 75) for the control glazed donuts, with 2n (double the sample size in each scenario) for the metformin donuts group. Fill in the blanks in the lawmaking chunk below to calculate the resulting ability for each of the three sample size scenarios.
pwr.2p2n.test(h = ES.h(p1 = __, p2 = __), n1 = seq(from = __, to = __, by = 25), n2 = seq(from = __, to = __, past = 50), sig.level = __, ability = Nada, alternative = "two.sided") You should get a power of 37.7% for the smallest n, 64.5% for n1=50/n2=100, and 81.4% for the largest n scenario.
add together chi foursquare
add correlation test
add anova
add together linear model
add note on guessing effect sizes - cohen small, medium, large
Explore More
You can explore other examples here in the official {pwr} vignette.
Power calculations for more complex endpoints and study designs can exist found in R packages listed in the Clinical Trials CRAN Task View here. Consider the packages {samplesize}, {TrialSize}, {clusterpower}, {CRTsize}, {cosa}, {PowerTOST}, {PowerUpR}, and which may be relevant for your item assay.
Two other helpful references are books:
Cohen, J. (1988). Statistical Power Analysis for the Behavioral Sciences (2nd ed.). LEA.
Ryan, T.P. (2013) Sample Size Determination and Power. Wiley.
How To Calculate Sample Size In R,
Source: https://bookdown.org/pdr_higgins/rmrwr/sample-size-calculations-with-pwr.html
Posted by: toddphrebre98.blogspot.com


0 Response to "How To Calculate Sample Size In R"
Post a Comment